Every day the world generates a vast amount of data in a variety of languages. Semantic networks, such as word co-occurrence networks (WCNs) can help overcome language barriers and analyze these data. Studies have shown that WCNs can accurately capture syntactical features of language by analyzing consecutive words in sentences, but thus far, no one has explored the relationships between distant words. Recently, researchers used an enhanced WCN to investigate just that.
There are nearly 7,000 different languages in the world and several quintillion bytes of data is generated in nearly all of them every day. This poses a serious problem for data analysis. Scholars have proposed complex network theory as a solution to this issue. One of the main types of semantic networks is the word co-occurrence network (WCN).
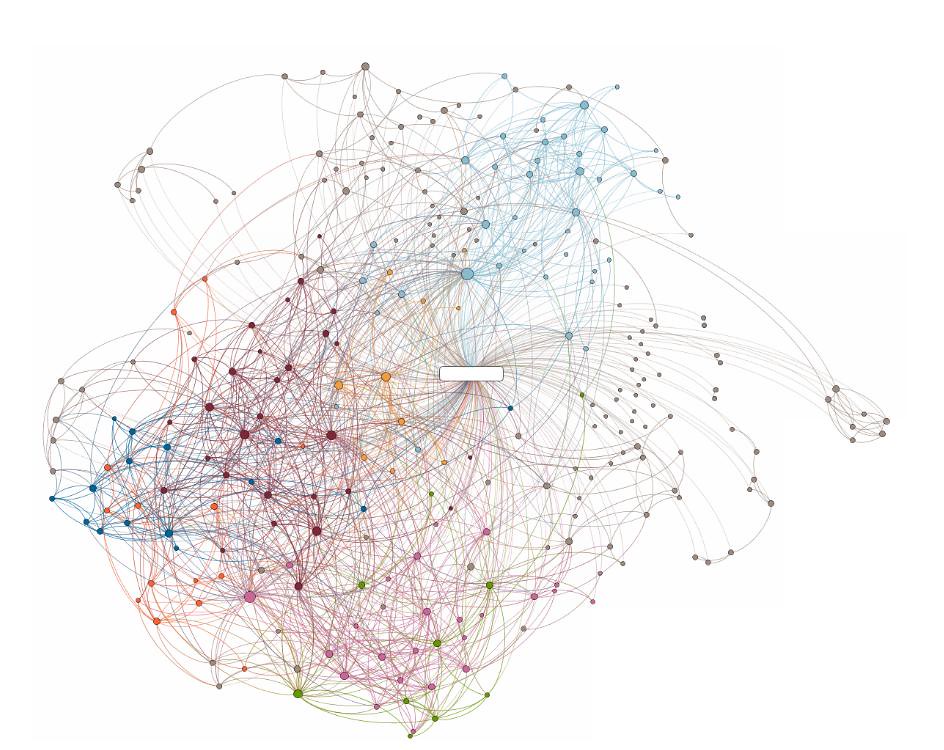
In a WCN, words form the vertices of the network (morphemes) and the edges between these vertices connect words on the basis of a string of words called an ‘n-gram.’ Here, n refers to the number of consecutive words in a sentence that are analysed at a time. Previous research has been limited to WCNs with a maximum n of two and have found that these WCNs can capture the characteristic features of multiple languages fairly well. But what is the relationship between distant words in sentences? Or, phrased differently, what happens when you increase the number of n beyond two?
To answer this question, a research team led by Prof. Tohru Ikeguchi from Tokyo University of Science, investigated the syntactic dependency relations in languages by using WCNs with increasing n. “We transformed well-known documents in eight languages into WCNs with n greater than or equal to two and found important features of each language in the WCNs,” says Professor Ikeguchi.
The team also consisted of Mr. Kihei Magishi and Prof. Tomoko Matsumoto of Tokyo University of Science and Prof. Yutaka Shimada of Saitama University. This study has been published in Nonlinear Theory and Its Applications, IEICE on April 1, 2022.
For their study, the research team transformed well-known works in eight different languages into WCNs. These works included a wide range of text data—the New Testament of the Christian Bible, the United Nations proceedings, the Paris agreement, and novels by different authors. These documents were chosen because they have been accurately translated into multiple languages, thereby allowing their faithful analysis. They then analysed the WCNs for a variety of n, up to n = 16.
“We found that the important features of each language appear in the networks with more than three co-occurrences, i.e., with n greater than or equal to three. We also saw that some of the network indices used to evaluate the structural features of the networks depend on the text data,” explains Prof. Ikeguchi.
The network indices that are dependent on the text data include the number of words and vertices, the density of the network, the triangle clustering coefficient and the square clustering coefficient. However, the research team also observed that some indices remained independent of the text data, such as the triangle clustering coefficient and the average shortest-path length, thereby enabling the description of the similarities and differences between languages.
Speaking of the long-term applications of the study, Prof. Ikeguchi says, “We are working towards the foundation of a new field of linguistics, mathematical linguistics. By deriving meta-grammar rules from mathematical commonalities and universality that appear in the grammatical functions of various languages, we will be able to establish a foundation for this field.”
The clarification of meta-grammar rules that do not depend on language will help realise the quantitative classification of language and help establish the factors that cause languages to diverge. The findings of this study constitute a major first step and make significant contributions to the understanding of the similarities and differences between languages.
***
Reference
Title of original paper: Investigation of the structural features of word co-occurrence networks with increasing numbers of connected words
Journal: Nonlinear Theory and Its Applications, IEICE
DOI: https://doi.org/10.1587/nolta.13.343
About The Tokyo University of Science
Tokyo University of Science (TUS) is a well-known and respected university, and the largest science-specialized private research university in Japan, with four campuses in central Tokyo and its suburbs and in Hokkaido. Established in 1881, the university has continually contributed to Japan’s development in science through inculcating the love for science in researchers, technicians, and educators.
With a mission of “Creating science and technology for the harmonious development of nature, human beings, and society”, TUS has undertaken a wide range of research from basic to applied science. TUS has embraced a multidisciplinary approach to research and undertaken intensive study in some of today’s most vital fields. TUS is a meritocracy where the best in science is recognized and nurtured. It is the only private university in Japan that has produced a Nobel Prize winner and the only private university in Asia to produce Nobel Prize winners within the natural sciences field.
Website: https://www.tus.ac.jp/en/mediarelations/
About Professor Tohru Ikeguchi from Tokyo University of Science
Tohru Ikeguchi received M.E. and Ph.D. degrees from Tokyo University of Science, Japan. After working for nearly a decade as Full Professor at Saitama University, Japan, he worked at Tokyo University of Science as Full Professor at the Department of Management Science from 2014 to 2016. Since 2016, he has been a Full Professor at the Department of Information and Computer Technology in Tokyo University of Science. His research interests include nonlinear time series analysis, computational neuroscience, application of chaotic dynamics to solving combinatorial optimization problems, and complex network theory. He has published over 230 papers and proceedings.
Funding information
This study was supported by JSPS KAKENHI Grant Numbers JP18K12701, JP20H00596, JP21H03514 and JP21H03508.