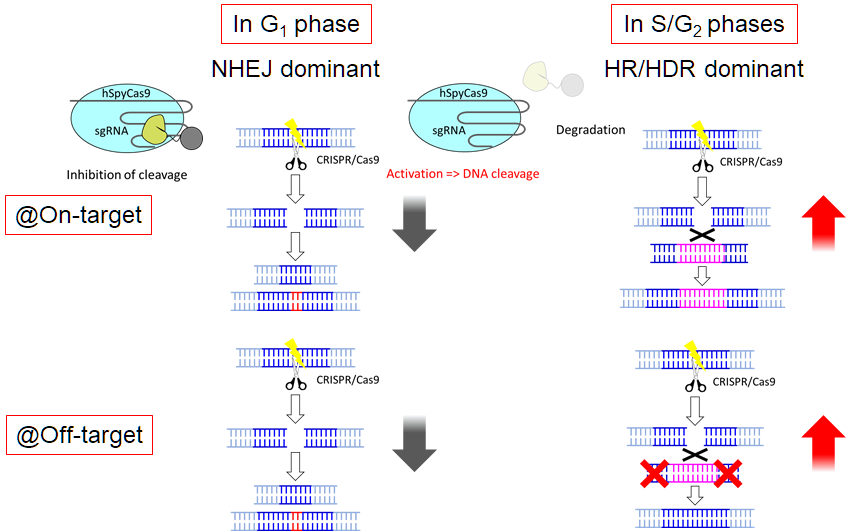
Scientists developed a promising fix to CRISPR-Cas9’s problem with unwanted genetic changes using a method that allows them to turn off gene-editing until it reaches key cell cycle phases where more accurate repairs are likely to happen.
Hiroshima University and Tokyo Medical and Dental University researchers published in
Communications Biology their study results which successfully demonstrated more precise gene-editing and suppressed unintended genetic mutations called off-target effects.
Although previous methods were developed that reported fewer off-target effects associated with the CRISPR technology, the researchers said these often exhibited lower editing efficiency.
“Our method is like hitting two birds with one stone. We can improve the preciseness of genome editing and suppression of off-target effects at the same time,” said
Wataru Nomura, study co-author and professor at HU’s
Graduate School of Biomedical and Health Sciences.
The newest method uses the anti-CRISPR protein AcrIIA4 which works like an “off switch” that stops SpyCas9’s genome editing activity. The researchers fused AcrIIA4 with the N terminal region of human Cdt1 — a gene that helps ensure DNA replication happens only once per cell division — intending to deactivate gene editing until S and G2, cell cycle phases when homology-directed repair (HDR) is dominant.
HDR is one of the two DNA repair processes used by organisms along with non-homologous end joining (NHEJ). Of the two, however, HDR is the preferred method as the repair relies on the existence of two chromosome copies in each cell. HDR’s use of the duplicate chromosome as a template for repair makes gene editing more precise as opposed to NHEJ which just tends to connect the broken ends of the DNA.
“The efficiency of HDR using AcrIIA4-Cdt1 was increased approximately by 4.0-fold compared to that using SpyCas9 alone,” the researchers said.
They found that at target or off-target site 1 (HCN1 gene), the mutation ratio decreased by 86.5%. The mutation ratio at off-target site 2 (MFAP1 gene) also decreased from 8.5% to 0.6%.
Nomura said they want to further improve the preciseness of the system so it could be used safely in the therapeutic field.
Image caption: This photo shows the possible mechanism of increased homology-directed repair efficiency and suppression of off-target effects by controlled activation of CRISPR-Cas9 depending on the cell cycle phase.
This story was first